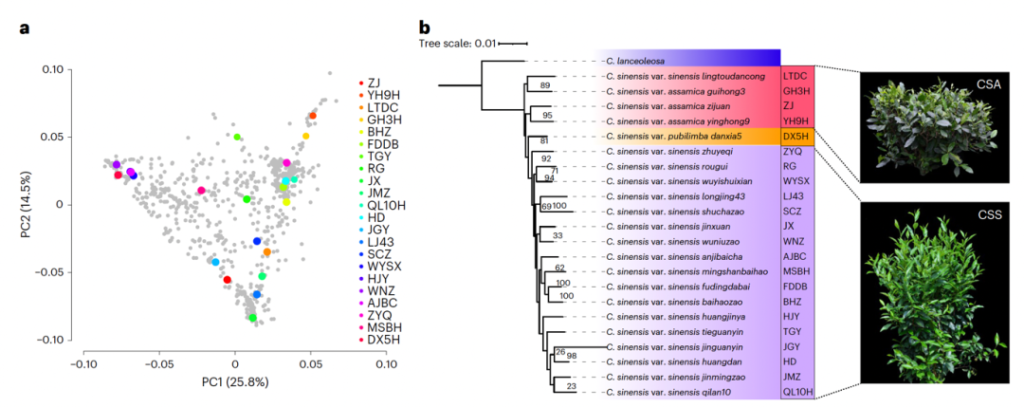
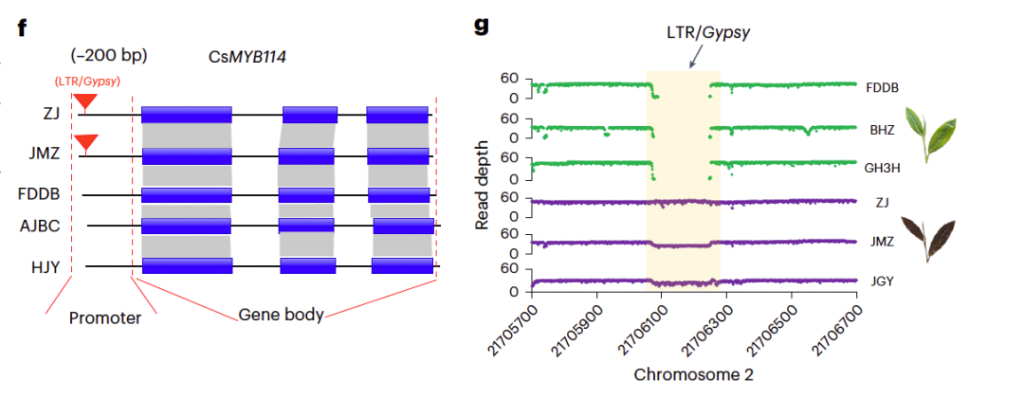
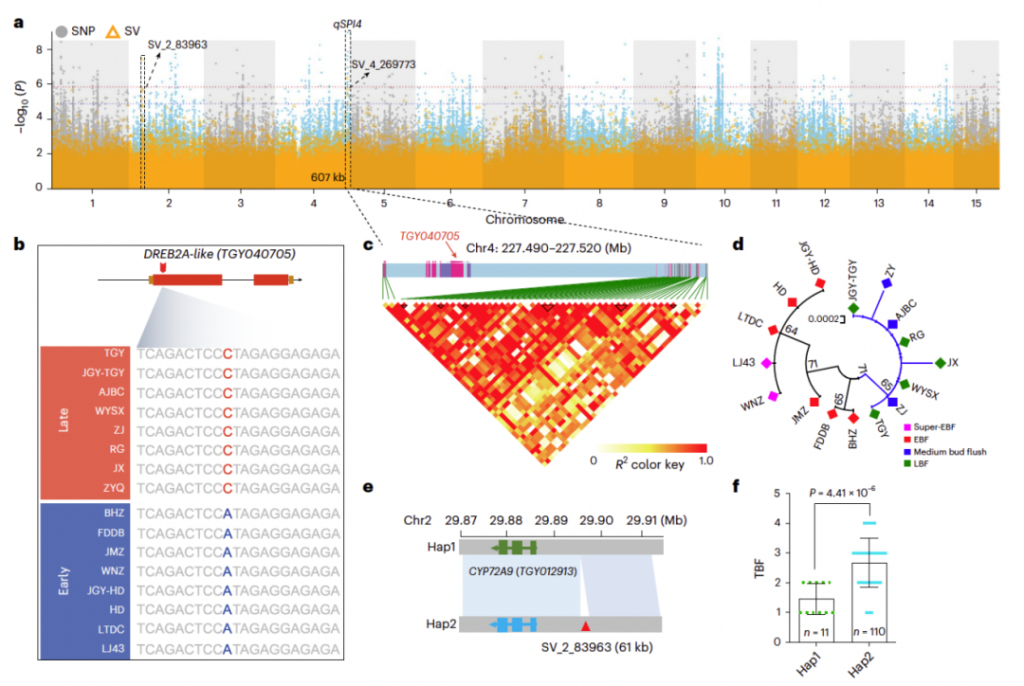
参考文献
1. Xia, E. et al. The Reference Genome of Tea Plant and Resequencing of 81 Diverse Accessions Provide Insights into Its Genome Evolution and Adaptation. Mol. Plant 13, 1013–1026 (2020).
2. Zhang, Q.-J. et al. The Chromosome-Level Reference Genome of Tea Tree Unveils Recent Bursts of Non-autonomous LTR Retrotransposons in Driving Genome Size Evolution. Mol. Plant 13, 935–938 (2020).
3. Wang, X. et al. Population sequencing enhances understanding of tea plant evolution. Nat. Commun. 11, 4447 (2020).
4. Zhang, W. et al. Genome assembly of wild tea tree DASZ reveals pedigree and selection history of tea varieties. Nat. Commun. 11, 3719 (2020).
5. Wang, P. et al. Genetic basis of high aroma and stress tolerance in the oolong tea cultivar genome. Hortic. Res. 8, 107 (2021).
6. Zhang, X. et al. Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant Camellia sinensis. Nat. Genet. 53, 1250–1259 (2021).
7. Tan, L. et al. Genetic analysis of the early bud flush trait of tea plants (Camellia sinensis) in the cultivar ‘Emei Wenchun’ and its open-pollinated offspring. Hortic. Res. 9, uhac086 (2022).
文章来源:植物生物技术Pbj